Here is an illustration of the Alignment API tutorial using the alignment server.
This tutorial has been designed for the Alignment API version 3.0 and updated for the Alignment API 4.0. Actual screendumps may vary since the interface has evolved and the functionalities have been extended.This tutorial can be used by locally launching an Alignment server or by using a publicly available Alignment server. We explain here how to install the alignment server.
Installing the Alignment server requires a jdbc-compliant relational database. Here we use MySQL. Drivers for Postgres are also available from the release.
Setting up the server requires to create a database. This is achieved through the following instructions:
Once this done, the server can be launched through command line by:
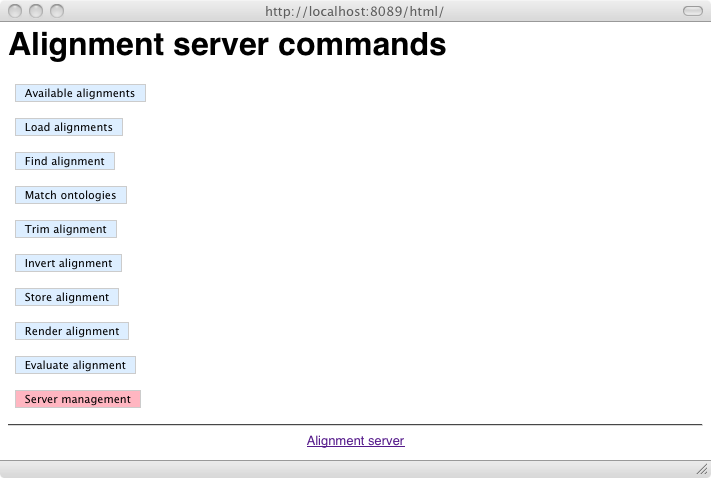
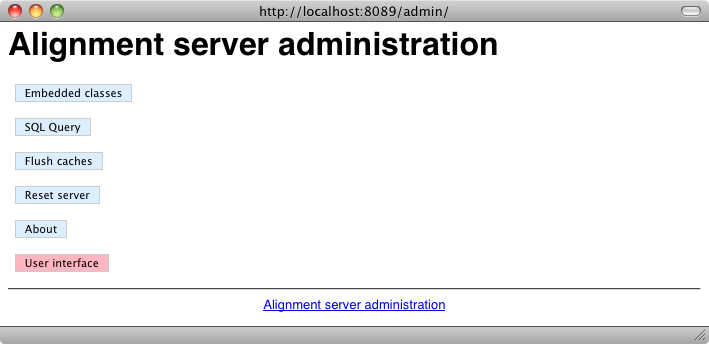
Once the server has been installed, it can be accessed using http://localhost:8089/html/. This provides access to two menus. The first one is the user menu:

The data is the same as that of the genuine tutorial. We will, however, use the versions which are available on the web at https://moex.gitlabpages.inria.fr/alignapi/tutorial/. They can be seen here:
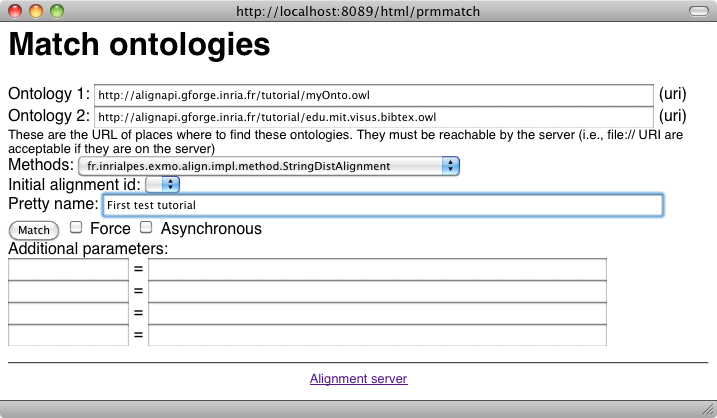
Your first matching task can be achieved by selecting the "Match ontology" button in the user menu:
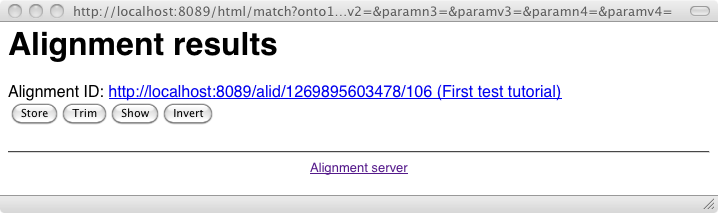
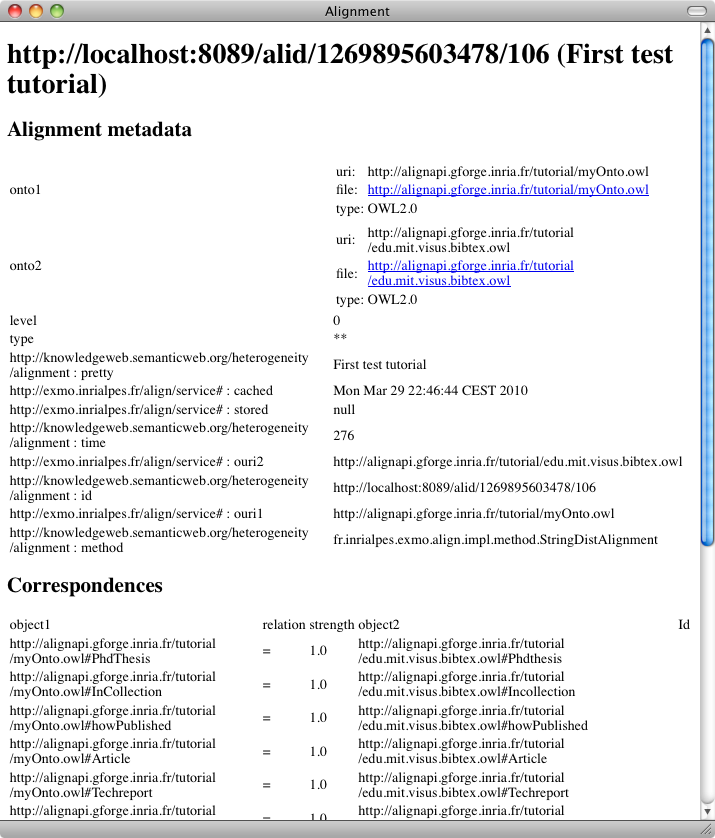
We will see later how to obtain the same result in different formats.
Other algorithms can be used by selecting other algorithm names or by passing different arguments to the same algorithm. The two algorithms used in the tutorial can be called as follows:
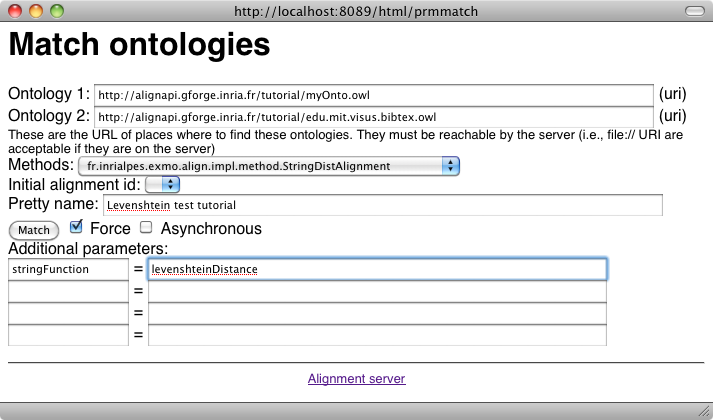
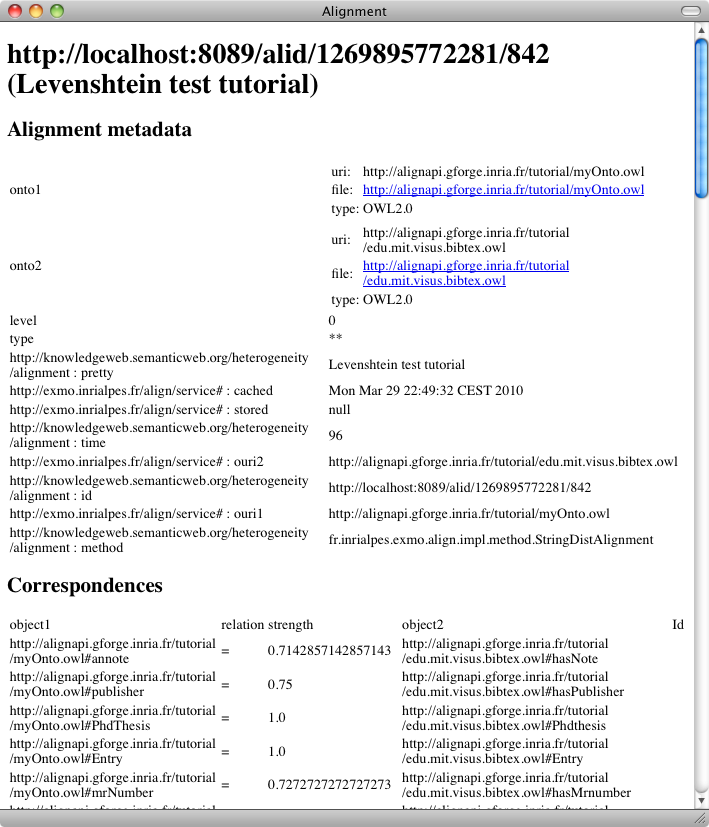
Look at the results: how are they different from before?
We can see that the correspondences now contain confidence factors different than 1.0, they also match strings which are not the same and indeed far more correspondences are available.
We can do the same with the other measure (the smoaDistance):
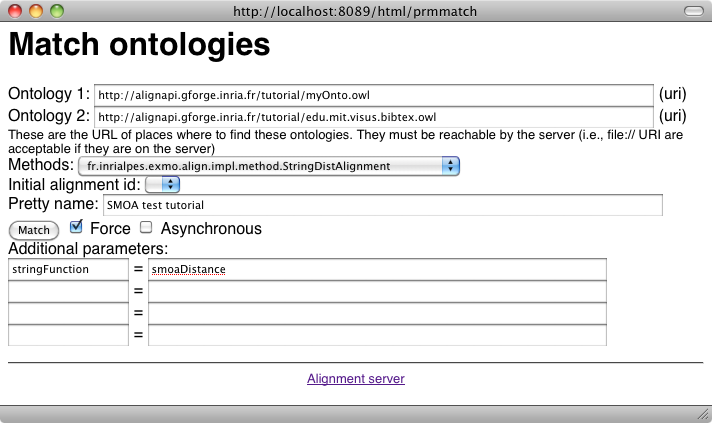
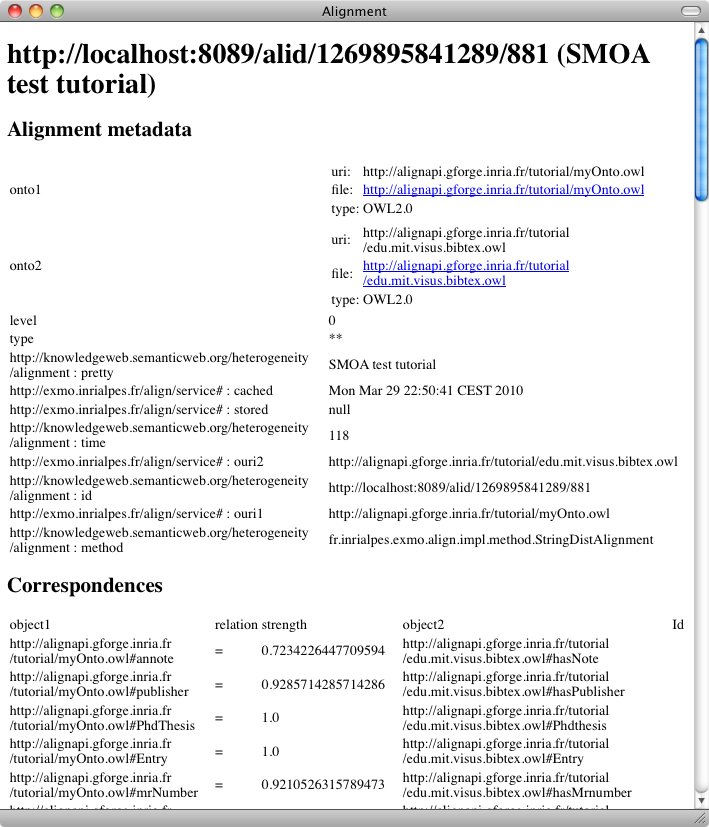
As can be seen there are some correspondences that do not really make sense. Fortunately, they also have very low confidence values. It is thus interesting to use a threshold for eliminating these values. Let's try a threshold of .33 over the alignment (with the -t switch):
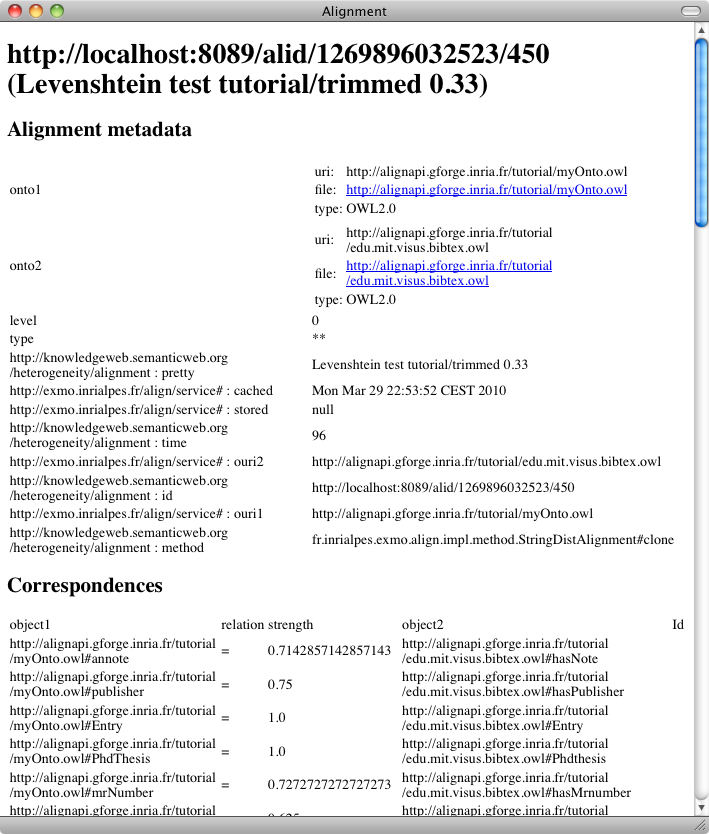
As expected we have suppressed some of these inaccurate correspondences. But did we also suppressed accurate ones?
This operation has contributed eliminating a number of innacurate correspondences like Journal-Conference or Composite-Conference. However, there remains some unaccurate correspondences like Institution-InCollection and Published-UnPublished!
More work: There is another switch (-T) in Procalign that specifies the way a threshold is applied (hard|perc|prop|best|span) the default being "hard". The curious reader can apply these and see the difference in results. How they work is explained in the Alignment API documentation.
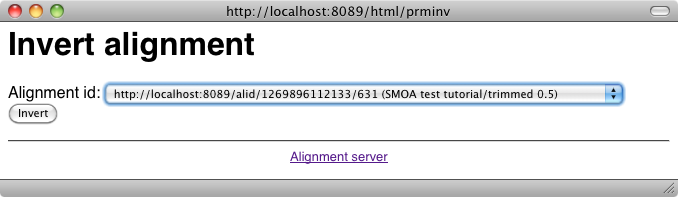
Other manipulations: It is possible to invert an alignment by using the invert button of the main menu or the one provided directly after results. This provides the following interface:
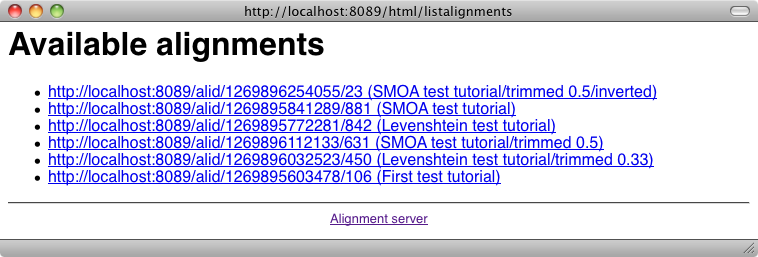
The list of alignments available from the server can by obtained from the main menu and gives:
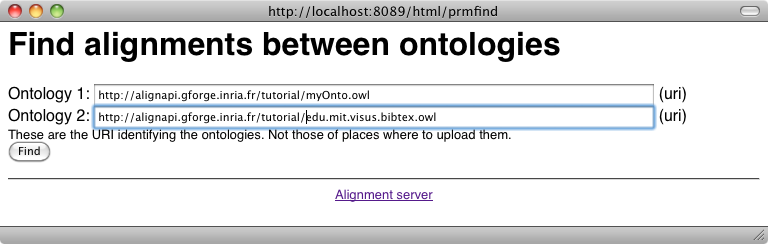
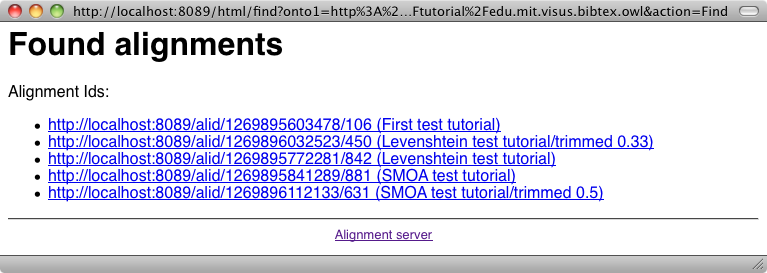
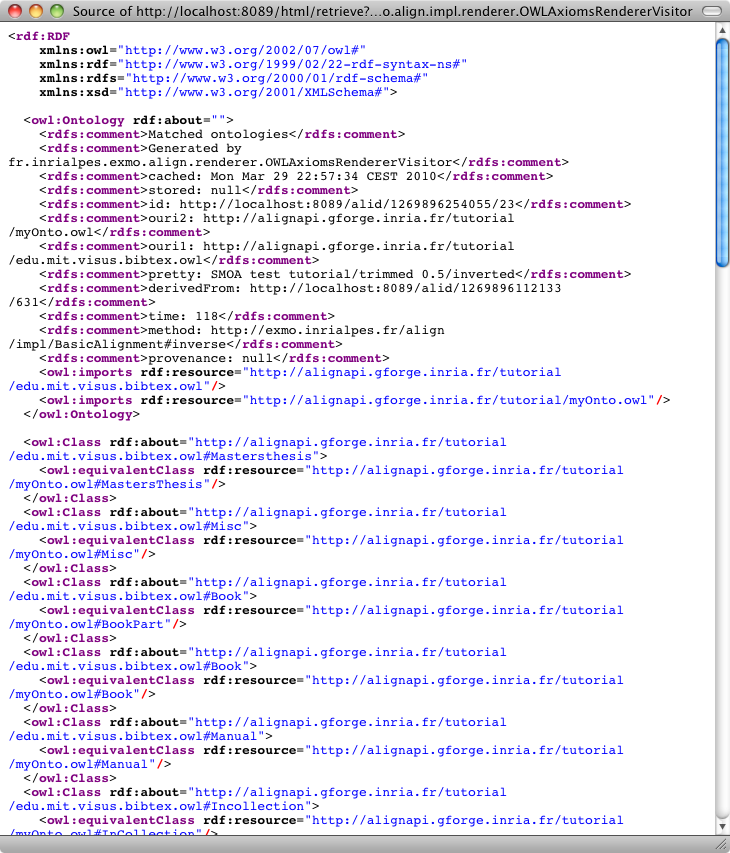
From the server, it is possible to generate all the formats available at the command line (depending on your browser, this may require to see the source of returned documents). This is achieved by using the "Render an alignment" button of the user menu:
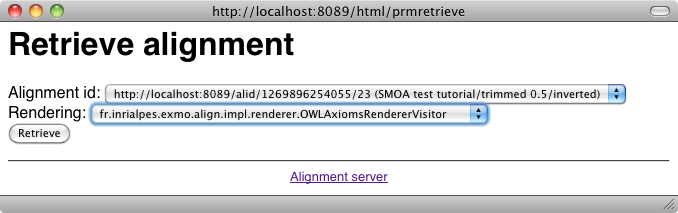
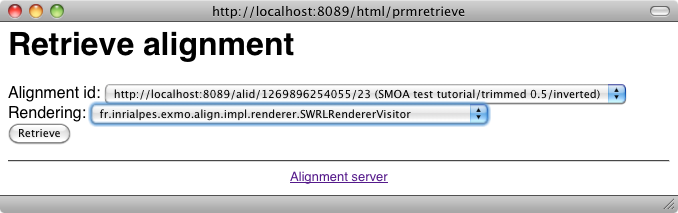
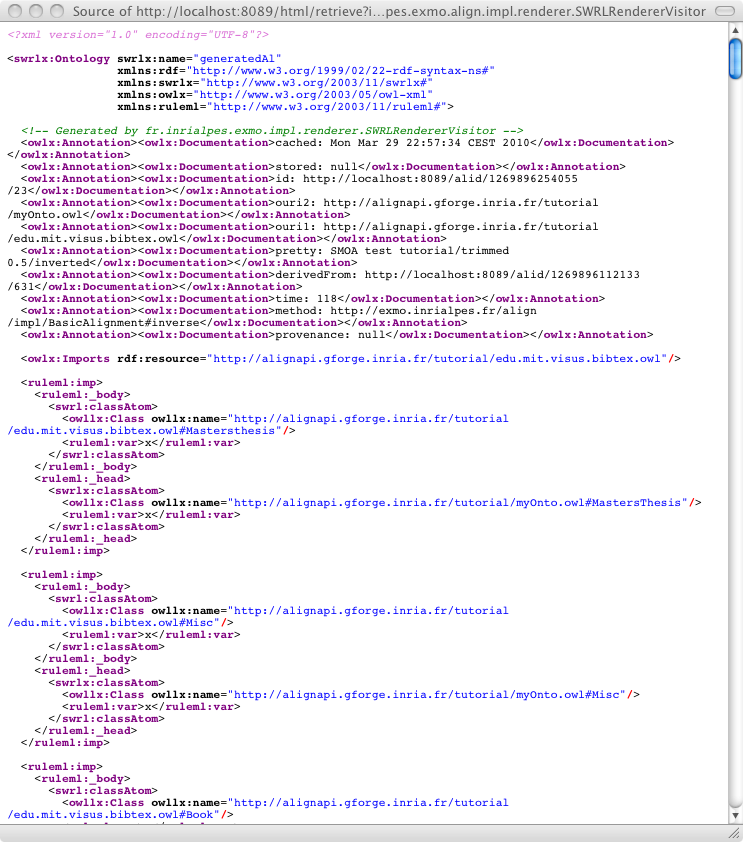
One can ask for the result as SWRL:
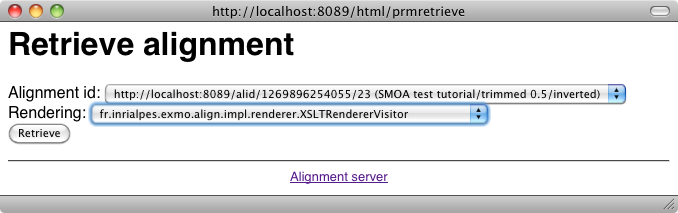
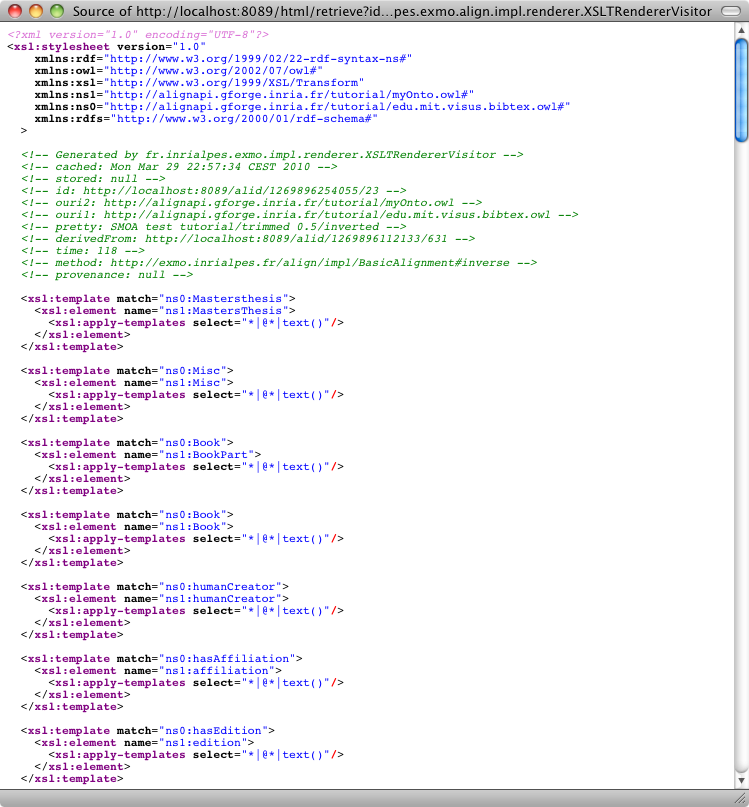
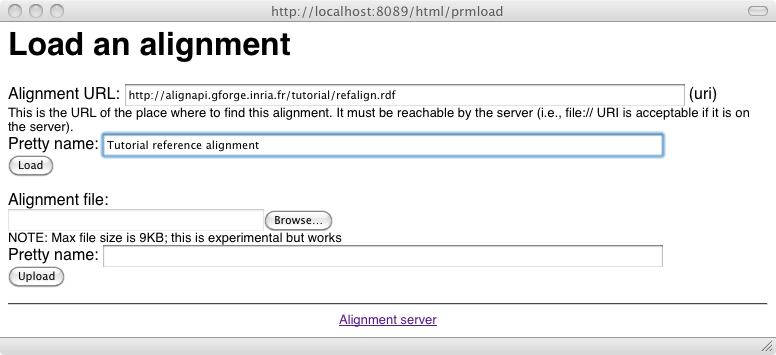
It is possible to load an alignment available somewhere on the web or on a disk through:
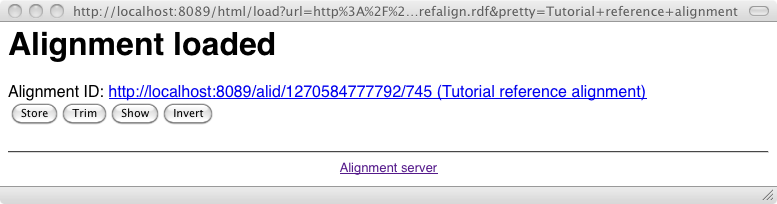
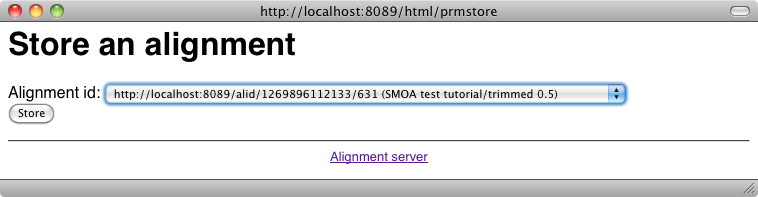
An alignment can be stored definitely on the server, i.e., its database through the Store button of the main menu which triggers the following interface:
An alignment can be compared to another (reference) one through the Diff Evaluator class: which returns three sets of correspondences (correct, incorrect and missing) :
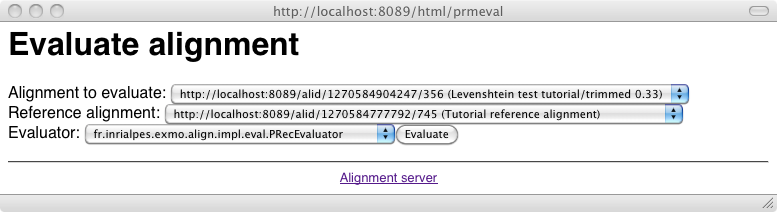
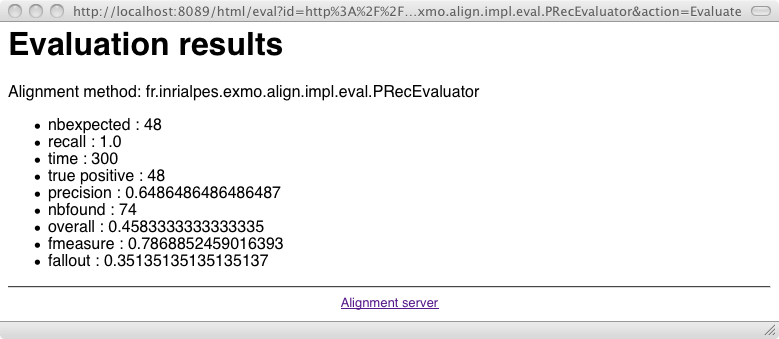
An alignment can be evaluated against a reference alignment through an Evaluator class:
More info: https://moex.gitlabpages.inria.fr/alignapi/tutorial/
https://moex.gitlabpages.inria.fr/alignapi/tutorial/tutorial1/server/